TargetATPsite
Service
- Web server: http://www.csbio.sjtu.edu.cn:8080/TargetATPsite/
Example
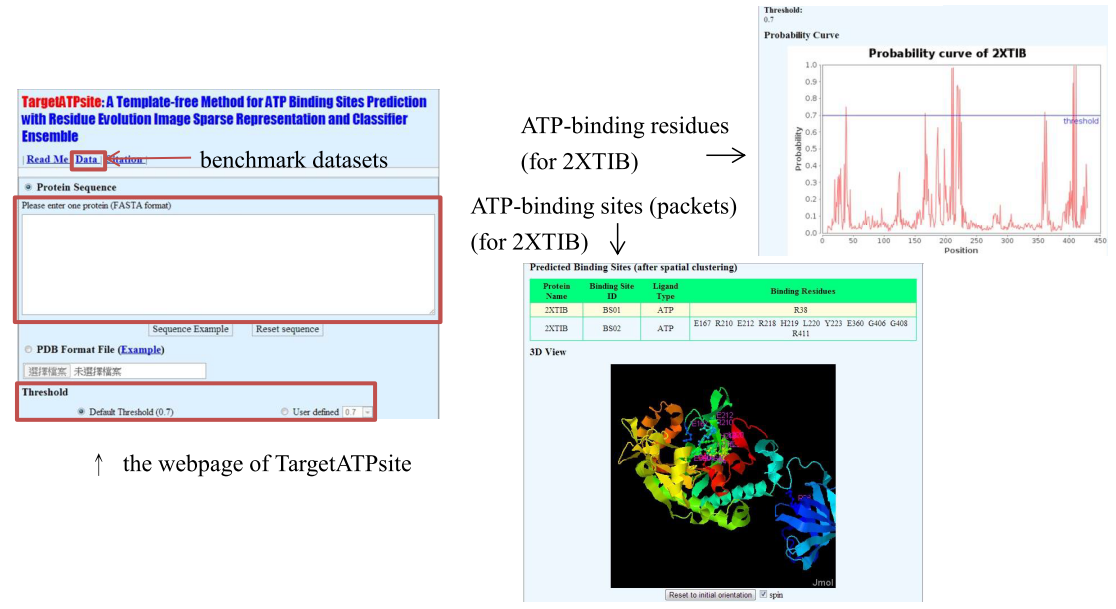
- Example : protein 2XTIB
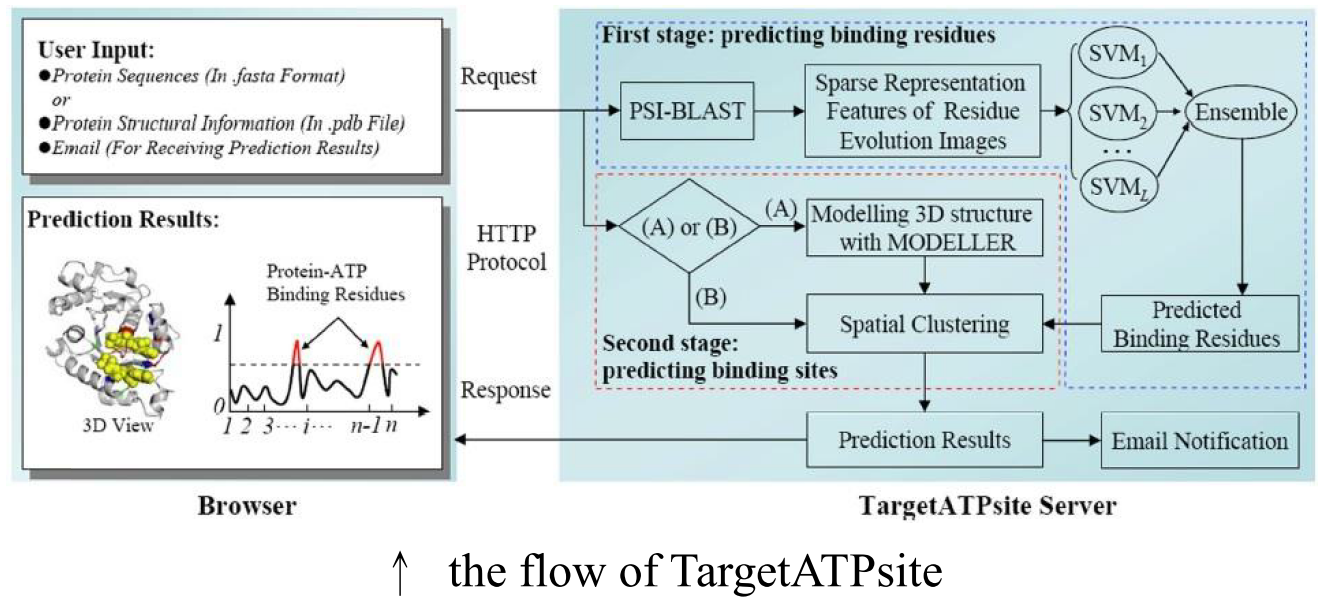
Classifier and Major method
an ensemble classifier: Adaptive boosting (Adaboost) with SVM is used to predict the binding residue of the primary sequence.
a spatial clustering algorithm is developed to find the binding sites (pockets) from the protein 3D structure
Dataset
- training dataset
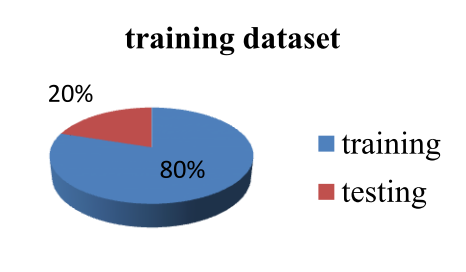
- Training protein sequences are random divided into 5 disjoint subsets, 4 subsets are for training and the remaining is for testing.
- 1$$^{st}$$ out of 2 databases, named ATP168: It consists of 168 non-redundant protein sequences which are selected from SuperSite encyclopedia by Chauhan et al. [1]
- 2$$^{nd}$$ out of 2 databases, name ATP227: It consists of 227 protein sequences from ref. [2] by Lukasz Kurgan et al.
Results
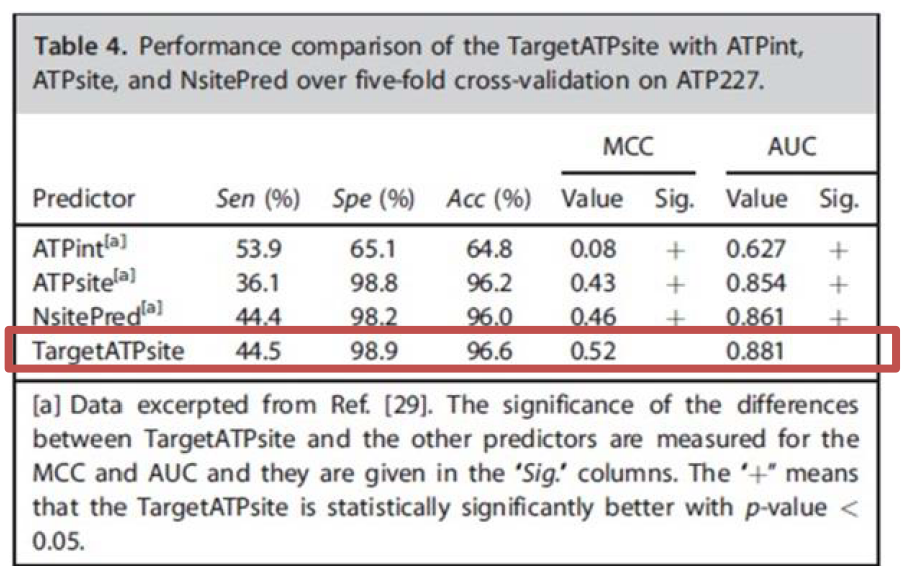
- The TargetATPsite perform significantly better than the existing predictors.
- MMC: Matthews correlation coefficients
- AUC: The area under the Receiver Operating Characteristic (ROC) curve and is related to threshold and indicates the prediction performance in positive
Independent dataset
- Independent dataset: A database containing 17 protein sequences as independent testing dataset is collected from ref. [2] by Lukasz Kurgan et al.
Reference
Jagat S Chauhan et al. (2009) Identification of ATP binding residues of a protein from its primary sequence. BMC Bioinformatics 10:434 doi:10.1186/1471-2105-10-434
Lukasz Kurgan et al. (2011) ATPsite: sequence-based prediction of ATP-binding residues. Proteome Science 9(Suppl 1):S4
Manning, G., Whyte, D.B., Martinez, R., Hunter, T. and Sudarsanam, S. (2002) The protein kinase complement of the human genome. Science 298, 1912-1934
Zhou FF, Xue Y, Chen GL, Yao X. (2004) GPS: a novel group-based phosphorylation predicting and scoring method. Biochem Biophys Res Commun 24;325(4):1443-8.
Shandar Ahmad, M. Michael Gromiha and Akinori Sarai (2004) Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. BIOINFORMATICS Vol. 20 no. 4, pages 477–486 DOI: 10.1093/bioinformatics/btg432
Dong-Jun Yu, Jun Hu, Yan Huang, Hong-Bin Shen et al. (2013) TargetATPsite: A Template-free Method for ATP-Binding Sites Prediction with Residue Evolution Image Sparse Representation and Classifier Ensemble. Journal of Computational Chemistry 34, 974–985