GPS2.0
Service
Java required
Web server : http://gps.biocuckoo.org/online.php
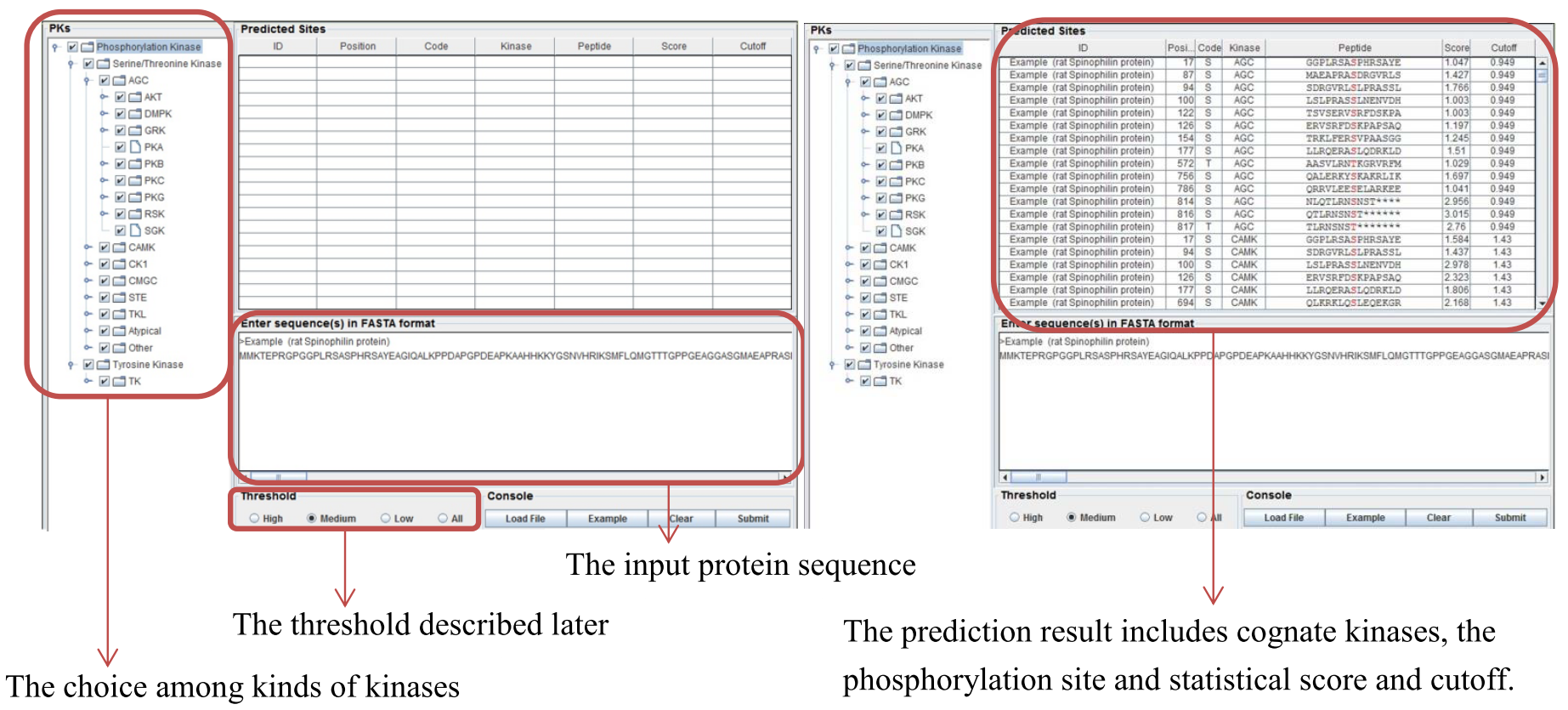
Example
- rat Spinophilin protein
Classifier and Major method
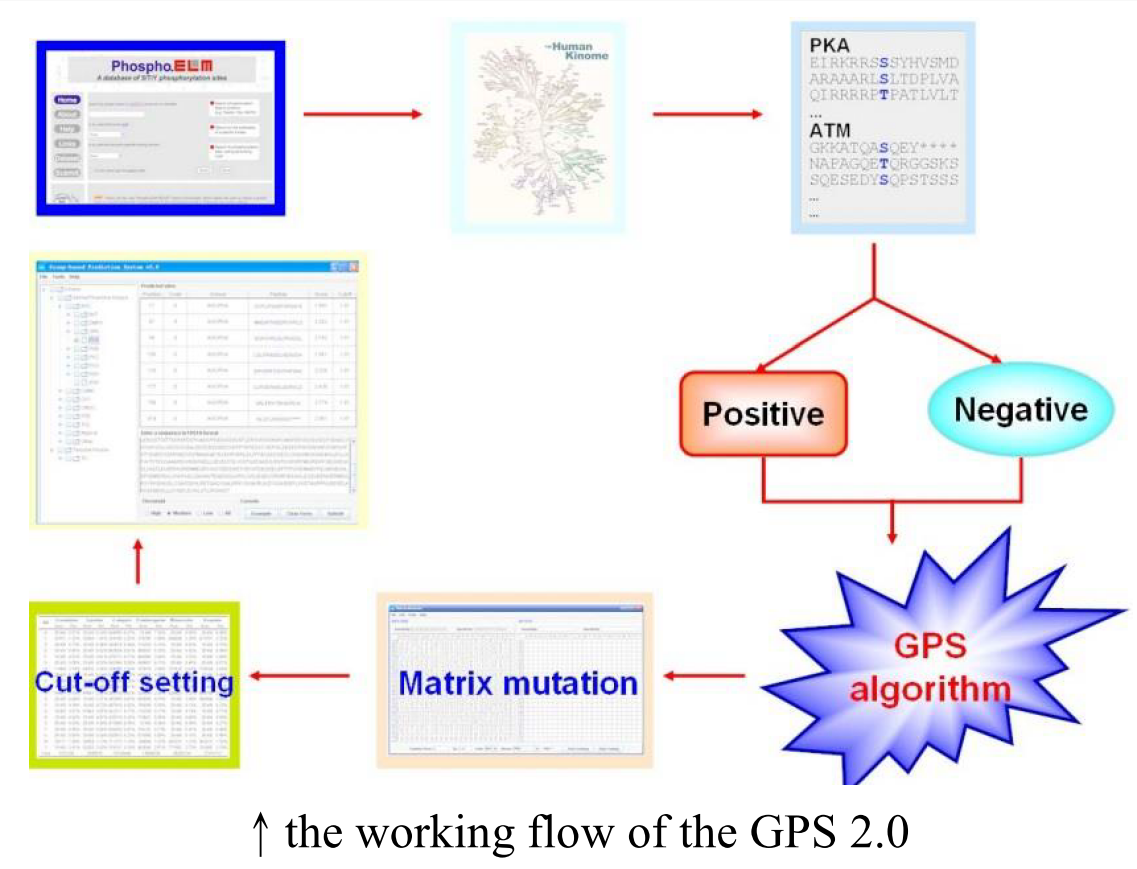
The first classifier grouping: The training protein sequences were classified into a specific hierarchical structure with four levels including group, family, subfamily and single PK. This method was developed by Manning et al. [3]
The second classifier GPS algorithm: calculate the score for each group and gain an original matrix. This method was developed by Zhou FF et al. [4]
The matrix is modified by the BLOSUM 62 for the robustness of the prediction.
The threshold with FPR:
| high | medium | low | |
|---|---|---|---|
| Serine/ Threonine | 2% | 6% | 10% |
| Tyrosine | 4% | 9% | 15% |
Dataset
The training dataset: It was derived from Phospho.ELM 6.0 [5], including 13,577 protein sequences (non-redundant).
The kinase information: There are 3,161 non-redundant sites correspond to their kinase information.
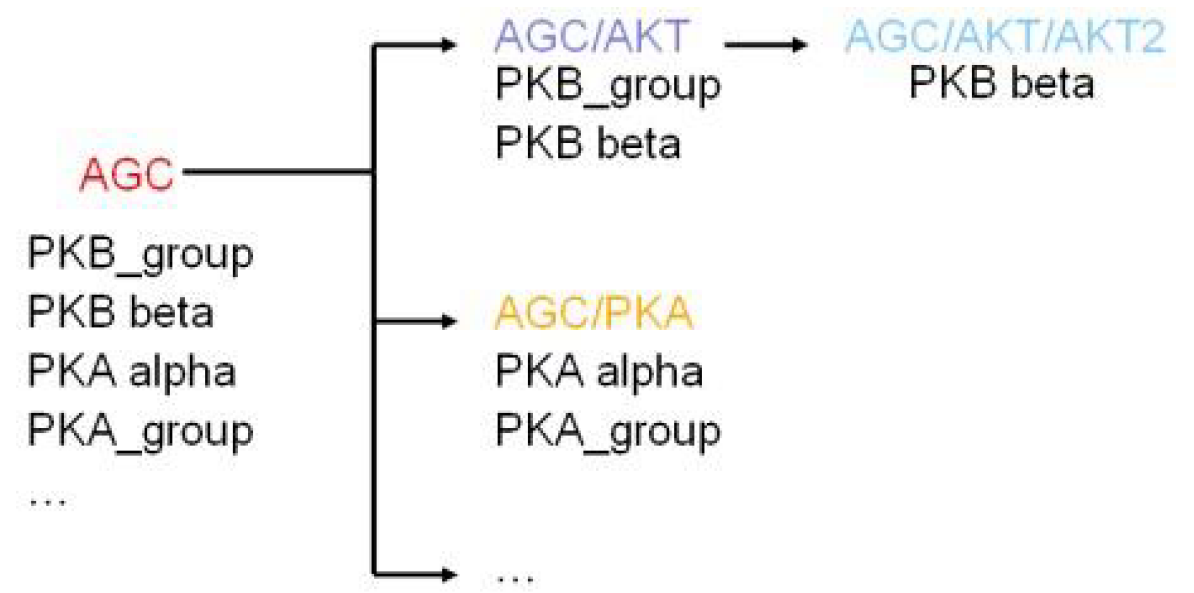
The training data could be re-used several times and included in different protein kinase clusters. Following the hierarchy of grouping the training data would be used in several times and narrowed down in order to specify the sub-group.
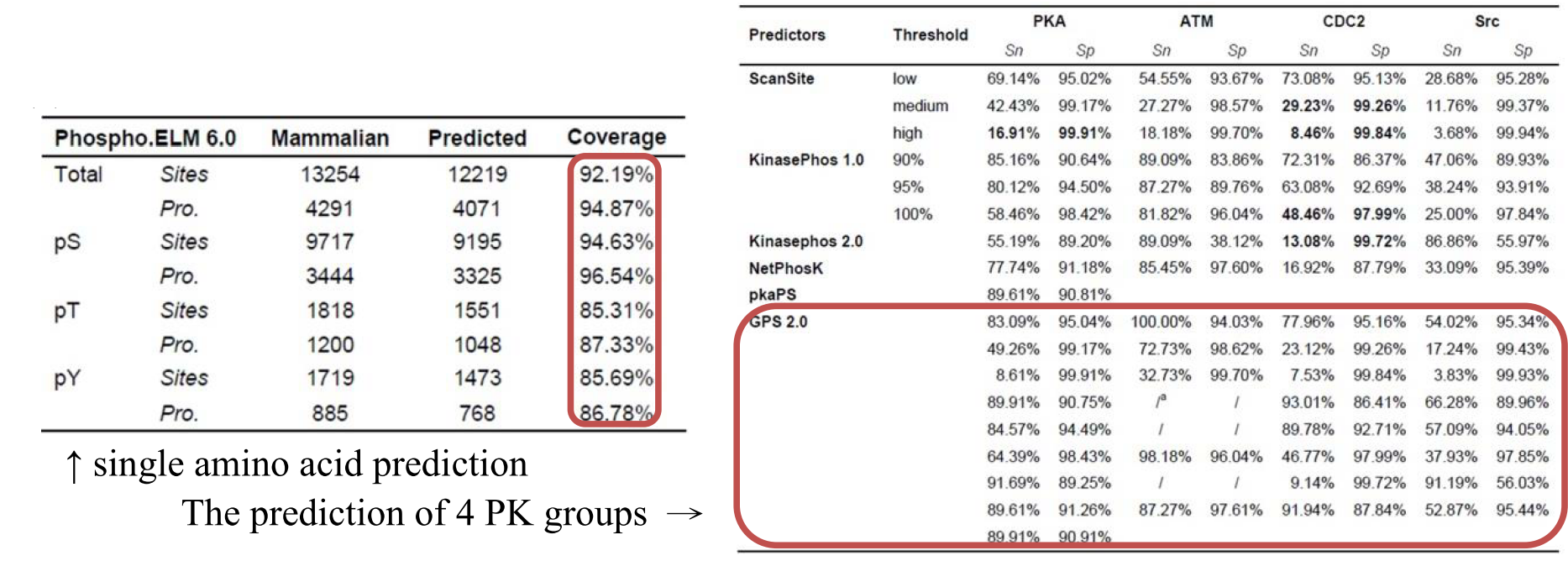
- The testing dataset: It is the same with the training dataset. The prediction performance was evaluated by self-consistency validation. And the Jack-knife validation and 4-, 6-, 8-, 10-fold cross-validation were extensively performed to evaluate the robustness and stability of the prediction system.
Results
- It is obvious that GPS 2.0 prediction performance is much better than other predictors.
Independent dataset
- There is no extensive data for testing in this paper.
Reference
Jagat S Chauhan et al. (2009) Identification of ATP binding residues of a protein from its primary sequence. BMC Bioinformatics 10:434 doi:10.1186/1471-2105-10-434
Lukasz Kurgan et al. (2011) ATPsite: sequence-based prediction of ATP-binding residues. Proteome Science 9(Suppl 1):S4
Manning, G., Whyte, D.B., Martinez, R., Hunter, T. and Sudarsanam, S. (2002) The protein kinase complement of the human genome. Science 298, 1912-1934
Zhou FF, Xue Y, Chen GL, Yao X. (2004) GPS: a novel group-based phosphorylation predicting and scoring method. Biochem Biophys Res Commun 24;325(4):1443-8.
Shandar Ahmad, M. Michael Gromiha and Akinori Sarai (2004) Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. BIOINFORMATICS Vol. 20 no. 4, pages 477–486 DOI: 10.1093/bioinformatics/btg432
Yu Xue, Jian Ren, Xinjiao Gao, Changjiang Jin, Longping Wen, and Xuebiao Yao (2008) GPS 2.0: Prediction of Kinase-Specific Phosphorylation Sites in Hierarchy. Molecular & Cellular Proteomics M700574-MCP200