scanProsite : a pattern recognition tool
Install & Operation
- Download the scanProsite program from ftp server and then uncompress the compressed file.
$ wget ftp://ftp.expasy.org/databases/prosite/ps_scan/ps_scan_linux_x86_elf.tar.gz
$ tar zxvf ps_scan_linux_x86_elf.tar.gz
Download the prosite database from ftp://ftp.expasy.org/databases/prosite/prosite.dat.
Change the path to access the necessary file: pfscan, psa2msa
- pfscan and psa2msa are located on line 690, 691

- Execute the perl script with several parameters and the following figure list several parameters for using the scanProsite.
$ perl ./ps_scan.pl -i ./ABL1.fa -o fasta > fasta_result.txt

- The following figure is the partial result from the above linux command.
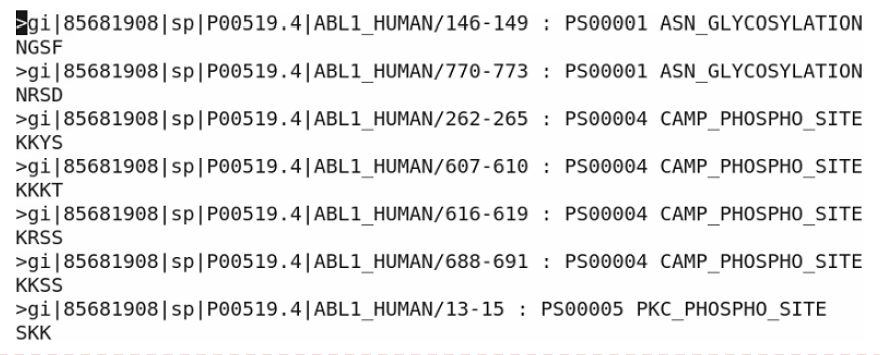
PROSITE Database
There are 72 sequences annotated by PROSITE patterns. The 7 out of 72 sequences are showed on the above figure. “/146-149” means the amino acid position (start-end).
There are about 10.263 (737 amino acids exist in 72 sequences) amino acid exists in a sequence annotated by PROSITE patterns. But this number could not represent the actual distribution of amino acids per sequence. There are several sequences with much more amino acids than the other ones.