iron imbalance and the iron responsive element
The introductions of ferritin
Ferritin is a ubiquitous intra-cellular protein. Its main function is storing iron, and is releasing iron in a regulation process . The amount of iron stored in ferritin reflects the total quantity of iron in a cell. Ferritin exists in almost all kinds of lives, including bacteria, higher plants and animals.
Ferritin serves as a primary protein storing iron in procaryotes and eukaryotes. In E. coli about 20% similarity to human ferritin is observed and each ferritin protein complex could store 4500 iron($$Fe^{3+}$$) ions[1].
Ferritin is a globular protein complex consisting of 24 subunits. It also play a role in possessing ferroxidase activity that makes ferrous($$Fe^{2+}$$) form covert to ferric($$Fe^{3+}$$) form.
Ferritin without iron is also named apoferritin.
The following 3-D structure of murine ferritin[1]. The central area is the place where iron stored.

An inherited disease affecting the iron-binding protein ferritin
Unlike the previous diseases discussed, such as sickle-cell anaemia[link]. There is a extremely rare disease called "hyper-ferritinaemia cataract syndrome".
There are two clinical symptoms of hyperferritin cataract syndrome, first is that the high level of ferritin protein in the blood and second is that lots of patient with cataracts in their eyes. The cataract causes the eye clouded.
The first case is described in 1995 at northern Italy from two families[2]. They shine under the glare and impaired visual acuity in childhood.
After, it was soon found that a mutation of ferritin mRNA causes this disease. Compare to the previous diseases discussed[link], such as sickle-cell anaemia and Huntington
s disease, the difference between hyperferritin cataract syndrome and them is the mutation site of mRNA. ** The mutation causing hyperferritin cataract syndrome is located at the regulation region in 5UTR of ferritin mRNA ** (the regulations of UTR refers to [link]).
Many proteins of iron metabolism are regulated at the level of translation
Free ferric is potientially toxic to the cell. Its ability is accepting electrons, meaning that if iron is free inside the cell, it can catalyze the conversion of hydrogen peroxide into free radicals. These free radicals may in turn affect others elements to cause a wide range of damages of cellular structures. To prevent these damages these irons are always captured by the specific proteins. These actions limit the harm to the cell. Beside the harm to cell, ferric iron may be needed in some biological processes. The concentration of ferric iron must be at suitable level. For almost organisms ferritin is the protein doing these actions, so the regulation to the gene expression of ferritin protein may carefully proceed.
The regulation of ferritin is involved in the translation of its mRNA. The major element regulating ferritin protein in its mRNA is several iron responsive elements(IREs) located in 5'UTR.
IREs correspond to two cytoplasmic proteins, iron responsive element binding protein I and II(IRP1 and IRP2), and they together regulate the production of ferritin protein.
In cells starved of iron IRPs may bind to IREs and this causes repression of translation activity of ferritin mRNA. This repression makes ferritin protein keep in a low concentration state. While iron is rich, IRPs tend not to bind IREs and this result in higher level of ferritin in order to maintain the concentration of iron. (refer to the following image[3])
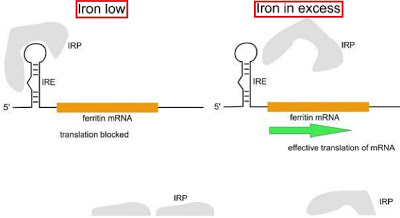
- the conceptual image of regulations that both IRPs and IREs operate.
- The major element, IREs, regulating the ferritin protein is located in 5'UTR of ferritin mRNA.
- When the concentration of iron is low, IRPs tend to bind IREs in order to repress the translation of ferritin mRNA and this make ferritin stay in a relatively low level of iron.
- When the level of iron in cell is relatively higher, IRPs tend to have lower affinity to IREs and this results in the repression of translation of ferritin mRNA.
- There is another protein carrying ferric iron ions, called transferrin. The transferrin receptor(Tfr) mRNA includes five copies of IREs located in its 3UTR. Two different proteins with the same mechanism to regulate the concentration of iron through the translation of its mRNA.
- the conceptual image of regulations that both IRPs and IREs operate.
Another protein carrying iron ions: tranferrin
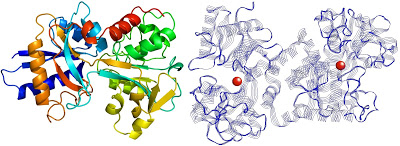
- The above image is 3D structual image of transferrin, the left side[4] and the right side[5] show where the iron ions store.
- Each transferrin carries two iron ions in ferric form($$Fe^{3+}$$).
- Beside ferritin transferrin also plays a role in transferring the iron ions in the blood.
- When transferrin delivers iron ions to a cell it binds to a transferrin receptor(Tfr) located in the surface of the cell.
- The transferrin receptor(Tfr) production is also involved in the translation of its mRNA and this mechanism is also regulated by five IREs located in 3'UTR of Tfr mRNA.
- Unlike the regulation of product of ferritin the regulation of transferrin expression is not involved in its translation process. The regulation of transferrin is controlled by IREs located in 3'UTR of transferrin mRNA.
- When the level of iron is reletively lower IRPs bind IREs located in 3'UTR of transferrin mRNA in order to stablize Tfr mRNA for gaining more iron ions outside. On the contrary, IRPs tend not to bind IREs when iron ions are abundant in the cell, as a result, the transferrin mRNA is stimulated to degrade in order to stop receiving the iron ions from out-cellular environment.
IREs are not only involved in mRNAs translation of ferritin and Tfr but also are present in other mRNAs whose products related to the iron metabolisms.
The IRE is part of RNA regulatory elements, in fact, there are more regulatory elements located in untranslated regions to regulate translation of mRNA(refer to "Regulation of mRNA translation by 5'- and 3'-UTR-binding factors"[link]). For example, bacterial riboswitches binding of a small molecule to the RNA structure and selenocysteine insertion sequence(SECIS) is responsible for incorporation of selenocystenine in proteins.
The structure of IRE
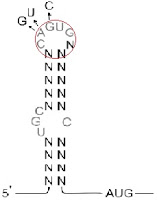
he structures of IREs is like a hairpin that consists of 26~30 nucleotids(refer to the above image[3]).
The partial loop sequence GAGUG is highly conserved in all IREs and is necessary for binding to the IRPs[6]. These GAGUG creat a bulge in the hairpin.
Analysis of one of the families showed a single mutation in the highly conserved loop sequence, CAGUG. Others families with same syndromes also show the mutations in the same sequence conserved. The mutations of these highly conserved sequences may cause hyperferritinaemia cataract syndrome[7]. All these mutations may interfere the binding of IRPs and IREs, as a result, these make the ferritin over produce.
Perl programming: Identify the IRE
IREs exist in ferritin, transferrin and others mRNAs related iron metabolisms. If there are some sequences similar with characters of IREs, these sequences might be regulated in the same way as ferritin and Tfr.
There are some barriers when finding the IREs-like sequences, such as:
- IREs contain a conserved sequence, GAGUG, but this sequence is too short to be a searching rule. Because such short sequence is ubiquitous in the whole genome.
- The sequence GAGUG is not located in the head or tail of the sequence and there are base-pairs between the head and the tail sequence.
The strategy finding IREs is quite simple through using a fixed size of the window. (refer to the following image)
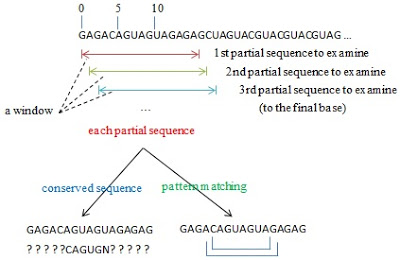
- Start from the beginning of the sequence, check a fixed window size of the sequence each time to identify whether it does exist the short conserved sequence in the partial sequence and whether there are base-pairing between the head and the tail in this partial sequence.
- If the partial sequence matches two above conditions, this partial sequence would be a IRE.
- If the partial sequence does not match both conditions, this partial sequence would be abandoned. The starting base of the whole sequence shifts to the next base and the partial sequence is re-produce and turn to the step 1 unless the end of the whole sequence.
- a simple implement in Perl programming as following
#!/usr/bin/perl -w
use strict;
# the whole sequence remained to check whether it is a IRE
my $wholeSeq = "GAGAGCAGUGGGGGUUUCCUGCUUCAACAGUGCUUGGACGGAACCCGGCGCUCGUU";
# subfunction: check whether two bases is complement
sub checkBasePair {
my ($base1, $base2) = @_;
if ( ($base1 eq 'G' and $base2 eq 'C')
or ($base1 eq 'G' and $base2 eq 'U')
or ($base1 eq 'A' and $base2 eq 'U')
or ($base1 eq 'C' and $base2 eq 'G')
or ($base1 eq 'U' and $base2 eq 'A')
or ($base1 eq 'U' and $base2 eq 'G')
)
{ return 1; }
return 0;
}
# subfunction: check whether the head and the tail sequence are each matching
sub findStemMatchStruct {
# as a return value
my $result = 1;
# get the value passed
my ($head,$tail) = @_;
# reverse the tail in order to compare easily
$tail = reverse($tail);
# each time get a base from each strands to compare further
for( my $j = 0; $j < 5 ;$j++ ) {
my $hBase = substr($head,$j,1);
my $tBase = substr($tail,$j,1);
# each time pass two bases to check the complement
# if there are one set of bases dismatching, result is modified to false(0)
if( checkBasePair($hBase,$tBase) == 0) {
$result = 0;
last; # equal to "break" in C-based languages
}
}
return $result;
}
# start to shift the window from each base of the sequence
for ( my $i = 0; $i < length($wholeSeq)-15; $i++ ){
# assume that the partial sequence consists of 16 base-pairs
my $testSeq = substr($wholeSeq, $i, 16);
# first rule: there is a conserved sequence
if ( substr($testSeq,5,5) eq "CAGUG" ) {
#seconde rule: several base-pairing between
#the head and the tail sequence
my $head = substr($testSeq,0,5);
my $tail = substr($testSeq,11,5);
#use two sub function to proceed pattern matching
if( findStemMatchStruct($head,$tail) ) {
my $pos = $i + 1;
print "match at pos: $pos\n";
print "$testSeq\n";
print "<----CAGUGN---->\n";
}
}
}
- the result from implementing this Perl script
match at pos: 23
UUCAACAGUGCUUGGA
<----CAGUGN---->
References
- Wikipedia (2013) Ferritin. http://en.wikipedia.org/wiki/Ferritin
- Girelli et al. (1995) Molecular basis for the recently decribed hereditary hyperferririne,ia-cataract syndrome: a mutation in the iron-responsive element of ferritin L-subunit gene(the 'Verona mutation'). Blood 86(11),4050-4053.
- Chapter 6。Genomics and Bioinformatics。Tore Samuelsson。Cambridge university press。July 2012。ISBN: 9781107401242。
- Wikipedia (2013) Transferrin. http://en.wikipedia.org/wiki/Transferrin
- Dr. N. Dennis Chasteen. (2013) Transferrin. http://pubpages.unh.edu/~ndc/
- Volz, K. (2008) The functional duality of iron regulatory protein 1. Curr Opin Struct Biol 18(1),106-111
- Kato. et al (2001) A mutation, in the iron-responsive element of H ferritin mRNA, causing autosomal dominant iron overload. Ann J Hum Gene 69(1), 191-197