Overlaps on sequence results from BLAST data
Deal with query results from BLAST(blastp)
- Consider the following example. The area surrounding by the green block is a part of query results from BLAST(blastp). The aim is to calculate the amount of genes in the forward and in the backward.
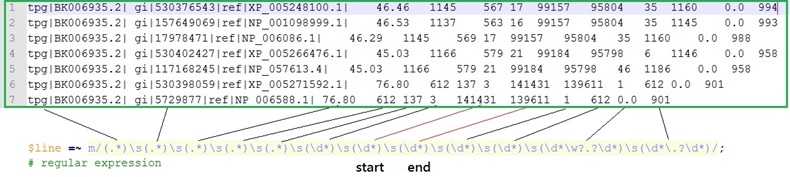
The first step is to extract the starting and end position of each query result. Further combine the several sequences into a much larger pattern based on two rules, that are overlaps and distance. (refer to the following image)
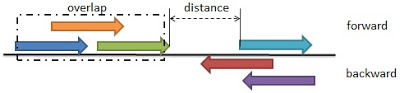
Overlaps mean intersections among several sequences that could be combined into one main sequence. Distance is referred as a space between two adjacent sequences. The execution condition takes both rules. The command used to execute the Perl script is the format as the following.
[~] $ perl ./combine.pl ./blast_out 10
- ./combine.pl: the script in Perl programming
- ./blast_out: the file input whose context is like the image above
- 10: the distance means two sequences are considered as one sequence in less than or equal to this value
Because the format of BLAST is the same and follows the strict rule it is powerful to use the regular expression to normalize each results . On the bottom of the first image above the regular expression designed for the query results is achieved by binding operator "~" and modifier "m".
- ' . ' : could be any characters without space-like ones, such as ' \t ', ' \n ', etc.
- \s : represents ' \t '
- \d : represents one digit number
- ' ? ' : a modifier represents the amount of existence, zero or one time
- ' * ' : another modifier represents the amount of existence, from zero to unlimited
Because of the regular expression it is easy to get the starting and end position with the helps of default memories saving each components in Perl programming . By extracting the default variable, $7 and $8, it is useful for receiving each query results. Position 7 and 8 are the counts of parentheses from the beginning of the regular expression.
After handling the regular expressions to the query results the following is to deal with the conditions in overlaps.
Concern about overlaps in several conditions
- There are several conditions needed to consider as following.
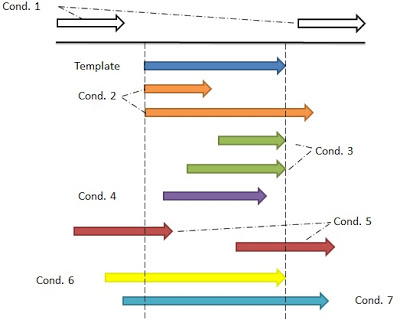
- Condition.1: no intersection with the template and two possible options, in the back or in the front of the sequence
- Condition.2: the sequences have the same starting position with the template but different end positions from the template
- Condition.3: the opposite of the condition.2
- Condition.4: the sequence is overlaped by the template
- Condition.5: either the starting position or the end position is involved in the part of the template
- Condition.6: the end position is the same with the template but the starting position is much smaller than the template
- Condition.7: the template is overlaped by the new sequence
These seven conditions are major parts to constitute the sub-function in Perl for checking the overlap between the new sequence(query result) and the template. Each condition has a part of Perl scripts that specific deal with it.
self-combination
- It is not enough to deal with the query result in the beginning. It is necessary to do self-combination and refer to the following image.
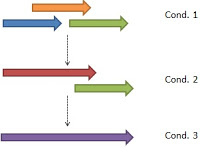
- Condition.1: the query results from the BLAST(blastp).
- Condition.2: After combinations among several sequences the hash saving the starting/end position would be like the result in the image.
- Condition.3: So it is necessary to combine elements themselves in the hash for the result in the image.
Implements
- The following is the implement for "Find the amount of genes in forward/backward sequence through overlaps on query results from BLAST data in Perl programming.".
#!/usr/bin/perl -w
use strict;
# Variables declaration to the file input and the value of gap distance input.
my $inputFile = "";
my $distance = 0;
for (my $i = 0 ; $i < 2 ; $i++) {
if ($i == 0) {
chomp($ARGV[0]);
$inputFile = $ARGV[0];
open(fin,$inputFile) or die ("File input error.\n");
}
elsif ($i == 1) {
chomp($ARGV[1]);
if(not $distance = $ARGV[1]) { die ("Lost the distance value.\n"); }
}
}
# Variables declaration stores the forward sequences, backward sequences and the flag detecting the overlap.
# The flag detecting the overlap is also used in the combination of internal sequences after.
my %forward = ();
my %backward = ();
my $flagInHash = 0;
# Show the content of the forward sequences and backward sequences saved in hash.
sub showHash {
# show forward
print "forward:\n";
foreach my $keyInHash ( sort{$a <=> $b} keys(%forward) ) {
print $keyInHash,"\t",$forward{$keyInHash},"\n";
}
# show backward
print "backward:\n";
foreach my $keyInHash ( sort{$a <=> $b} keys(%backward) ) {
print $keyInHash,"\t",$backward{$keyInHash},"\n";
}
}
# This sub-function tries to combine sequences that overlap another sequence into one.
sub overlap {
my ($start, $end, $option) = @_;
# Pass the value, not reference.
my %combineHash;
if($option == 1) { %combineHash = %forward; }
elsif($option == 2) { %combineHash = %backward; }
# $flagInHash == 0 means starting searches or no overlap in the hash.
# $flagInHash == -1 means overlap happened. Two sequences were combined already.
# Besides that, -1 means a overlap happens and is used in the internal overlap checks.
$flagInHash = 0;
foreach my $element (keys(%combineHash)) {
# The starting position of the new sequence input is the same with a sequence stored in the hash.
# If the end position of the new sequence input is smaller to the sequence stored in the hash, nothing to do.
# On the contrary, it is necessary to replace the value related the same key.
# If the end position is equal to the value stored in the hash it must continue to the next element in the hash.
# If it is not continue internal overlap checking could be wrong.
if($start == $element) {
if ($combineHash{$start} < $end) { $combineHash{$start} = $end; }
elsif ($combineHash{$start} == $end) { next; } # used in the internal overlap checking.
$flagInHash = -1; # already changed
last;
}
# The starting position of the new sequence is not the same with anyone in the hash.
# Instead the end position of the new sequence gets a hit in one of the hash.
elsif($end == $combineHash{$element}) {
# the head is larger: the hash item overlaps the new sequence and nothing is changed
if($start > $element) {
$flagInHash = -1; # already changed
last;
}
else {
# the head is smaller: new sequence overlaps a sequence in the hash
# the item in hash is deleted and new sequence is created and saved into the hash
delete($combineHash{$element});
$combineHash{$start} = $end;
$flagInHash = -1; # already changed
last;
}
}
# the sequence input is totally overlaped by the item in the hash and nothing is changed
elsif($element < $start and $end < $combineHash{$element}) {
$flagInHash = -1; # already changed
last;
}
# the end position of the sequence input is larger than the sequence in hash, but starting position is inside the item
# it should notice the boundary of the item in hash and the equal condition must be considered
elsif($element < $start and $start <= $combineHash{$element} and $combineHash{$element} < $end) {
$combineHash{$element} = $end; # elongation
$flagInHash = -1; # already changed
last;
}
# the end position of new sequence between $element and its hash value,
# but the starting position of the new sequence is smaller than the item in hash
# # it should notice the boundary of the item in hash and the equal condition must be considered
elsif($start < $element and $element <= $end and $end < $combineHash{$element}) {
# elongation: create a new item, start from the new sequence and end to the old sequence(item) in hash
# the smaller item in hash sould be deleted
$combineHash{$start} = $combineHash{$element};
delete($combineHash{$element});
$flagInHash = -1; # already changed
last;
}
# new sequence completely overlaps the item in hash
elsif($start < $element and $combineHash{$element} < $end) {
# delete the item in hash and create a new item to save into the hash
delete($combineHash{$element});
$combineHash{$start} = $end;
$flagInHash = -1; # already changed
last;
}
}
# When no such overlap mentioned before it should be created a new item based on the new sequence and store it into the hash.
if ($flagInHash == 0) {
$combineHash{$start} = $end;
}
# It should save back into the global variable because of the method of passing the value.
if($option == 1) { %forward = %combineHash; }
elsif($option == 2) { %backward = %combineHash; }
}
# selfCombine is designed for internal overlap checks.
sub selfCombine {
my $deleteCheck = 0;
my $stopSelfCombine = 1; # used as a flag to confirm that the self combination is finished
while ($stopSelfCombine) {
# while loop is used to confirmed that the sequences are completely combined through variable $flagInHash
$stopSelfCombine = 0;
foreach my $element (keys(%forward)) {
# because keys(%forward) may return a list stored in memory, e.g. memA, internal overlap checking may delete
# a pair and the key of this pair may previous be saved into memA. However, check over the internal overlap and return
# the control to the loop, loop may get the next value from memA and this may go wrong cause of deleting the value.
# Use another foreach each time before the loop getting the value is to check whether the key previous stored is still
# in the hash.
$deleteCheck = 0;
foreach my $keyIter (keys(%forward)) {
if($element == $keyIter) { $deleteCheck = 1; last; }
}
if($deleteCheck == 1) {
overlap($element, $forward{$element},1);
# if there is still a replacement it would repeat one more loop to check that the overlap is finished.
if($flagInHash == -1) { $stopSelfCombine = 1; }
}
}
}
$stopSelfCombine = 1;
while ($stopSelfCombine) {
$stopSelfCombine = 0;
foreach my $element (keys(%backward)) {
$deleteCheck = 0;
foreach my $keyIter (keys(%backward)) {
if($element == $keyIter) { $deleteCheck = 1; last; }
}
if($deleteCheck == 1) {
overlap($element, $backward{$element},2);
if($flagInHash == -1) { $stopSelfCombine = 1; }
}
}
}
}
# distanceCombine is designed for consideration the distance between two sequences in the hash.
sub distanceCombine {
# first each sequence adds the value of the distance and then undergos the sequences database self combinations
# the minus of the starting position in backward sequence is equal to the plus of the starting position in the forward sequence
foreach my $key (keys(%forward)) { $forward{$key} += $distance; }
foreach my $key (keys(%backward)) { $backward{$key} += $distance; }
# it is necessary to do the self-combination again
selfCombine();
}
# This foreach loop deals with the data input in BLAST format 6.
foreach my $line () {
# remove new line
chomp($line);
# regular expression
$line =~ m/(.*)\s(.*)\s(.*)\s(.*)\s(.*)\s(\d*)\s(\d*)\s(\d*)\s(\d*)\s(\d*)\s(\d*\w?.?\d*)\s(\d*\.?\d*)/;
# forward sequence
if ($7 < $8) {
overlap($7,$8,1);
}
# backward sequence
else {
# the direction is not a problem when it comes to talk about overlap
overlap($8,$7,2);
}
}
# internal overlap checks
selfCombine();
# combine with a distance and sequence overlap checks in hash
distanceCombine();
# output the data
#showHash();
# show the final result
print scalar(keys(%forward)),"\n",scalar(keys(%backward)),"\n";
# file closed
close(fin);