PCR : Amplifying DNA
Background
Consider the following conditions:
- you have to produce DNA in a larger quantity,
- a longer DNA molecule, for example, a whole human chromatin, is hard to work with and
- there is only a smaller region of DNA we interest
There are two common solutions,
- first one is a way of cloning, a smaller DNA fragment is introduced into a circular plasmid which allows the bacteria to replicate in the cell. Through the purification of plasmids and cutting its upon restriction enzymes, we could get the smaller DNA fragement we insterest.
- The other is ploymerase chain reaction(PCR) invented by Kary Mullis in 1984.
What is PCR?
As mentioned before,
- PCR is useful for amplification a shorter region of DNA and for producing a distinct region of DNA in a large quantity.
- PCR could be implemented in a test cube with the help of DNA polymerase(a kind of enzyme).
- One of the advantages of PCR is that the DNA product is relatively pure.
How do PCR operations proceed? Before operations of PCR, there is a assumption that you know a piece of sequence towards the ends of region you want to amplify. The construction of primers is constructed on these regional sequence you intend to amplify.
Three cycles during the PCR processes,
- Heating: the first step in PCR is that separating the sequence you want to amplify with the help of heating.
- Annealing: the second step is that primers constructed associate with DNA strands on the basis of sequence complementarity. There are two primers which associate with sense and anti-sense strand of target DNA sequence.
- Extension: DNA synthesis occurs from the location of primers, meaning extending from the primers.
We could perform the multiple cycles in the same test tube by adjusting the temperature. A high temperature is needed in the first step of PCR processes, so a thermo-stable DNA polymerase is necessarily taken. Such enzyme may be extracted from bacteria lives at the high temperature region, i.e. hot springs.
In each cycle, the amount of DNA is double ideally. For example, the amount of DNA has increased in theory by factor of $$2^{30}$$.(~= $$10^{9}$$)
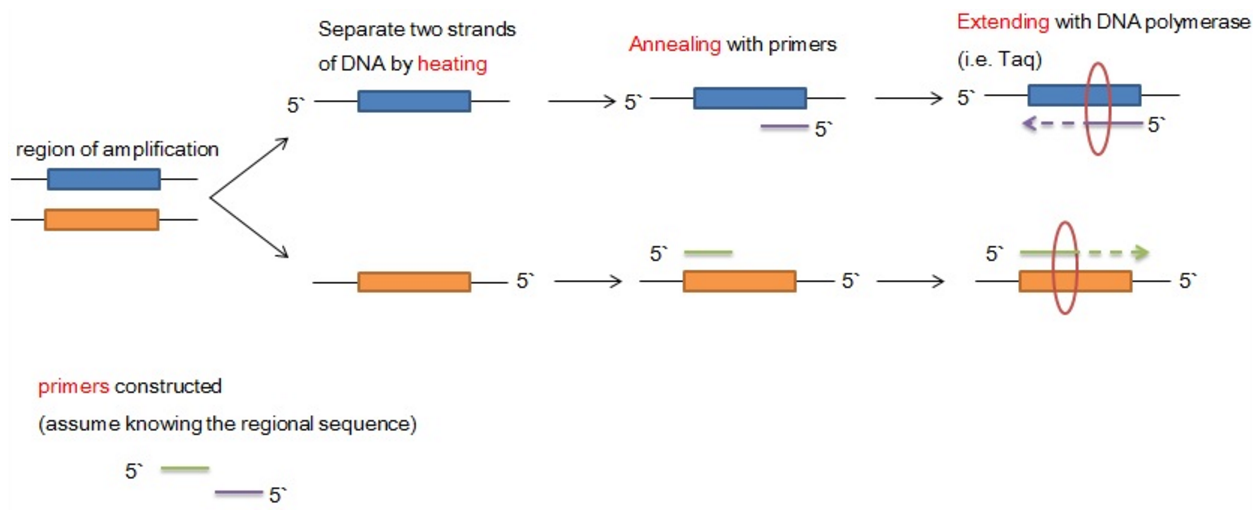
- The region of DNA double strands is amplified.
- Short oligo-nucleotide sequences known as primers constructed before the PCR.
- The DNA is first heated to about $$95^{o}C$$ to allow double strands of DNA to seperate.
- The temperature is then lower to $$50^{o}C - 65^{o}C$$ to allow annealing of two different primers to the DNA strands after the primer associates with 3' end of the DNA strand.
- The third step is extending from DNA primers in 5' to 3' direction.
The replicaion of DNA is semi-conserved process. PCR is also extremely sensitive. This sensitivity is useful whenever the amount of DNA in a sample is restricted.
PCR Applications
PCR is used in diagnosis of viral disease such as HIV, rabies, and in diagonsis of becterial indections like tuberculosis which pathgens grows slowly.
PCR is used to monitor cancer therapy as to detect cancer-specific chromosomal abnormality.
PCR is used to analyse ancient DNA which could be thousands of years old.
PCR is used to re-construct a portion of the whole genome of a species.
PCR primer design
- primer length should be in the range 18 ~ 22 nucleotides
- melting temperature should be 95 (celsius); the temperature at which half of the DNA duplex will dissociate to single-stranded entities.
- GC content should be 40 ~ 60 %
- sequences with stable secondary structure should be avoided like head-tail sequence matching problem
- repeated should be avoided
- long runs of single bases should be avoided
- primer should be as more as specific to target DNA (check with BLAST)
design primers of an unknown organism
- design a primer which is highly conserved in sequence, like rRNA gene.
- the species of interest has close relatives where the sequence is known.
design primers of protein-coding gene without sequence data
- Assume that protein-coding sequence is known. An example is following,
Gly-Val-Thr-Lys-Trp-Lys-Met
We could reverse translation of the amino acid sequence into RNA sequence. Of course there are some problems, e.g. serine is translated from UCU, UCC, UCA, UCG, AGU, AGC. As a result, we may make use of mixture primers where all possible primers are represented, that is, we can infer all possible RNA sequences with the help of genetic code table,
protein: Gly - Val - Thr - Lys - Trp - Lys - Met
DNA: GGN - GTN - ACN - AAR - TGG - AAR - TAG
N: A, T, C, G
R: A, G
There are 4 x 4 x 4 x 2 x 2 = 256 possible sequences for designing primers. Therefore we must avoid too many uncertain oligo-nucleotides in order to operate PCR.
Design primers of a protein-coding sequence in Perl
use strict;
# hash table, 每個 amino acid hash 到不同的 tri-nucleotide sequence
my %reverse_code = ('L', 'YUN', 'F', 'UUY', 'I', 'AUH', 'M', 'AUG',
'V', 'GUN', 'S', 'WSN', 'P', 'CCN', 'T', 'ACN',
'A', 'GCN', 'G', 'GGN', 'Y', 'UAY', 'H', 'CAY',
'Q', 'CAR', 'N', 'AAY', 'K', 'AAR', 'D', 'GAY',
'E', 'GAR', 'C', 'UGY', 'W', 'UGG', 'R', 'MGN');
# reading protein sequences from a file and link all sequence without '\n'
my $proteinSeq = "";
open(fin,"brca1.pep") or die("Input file error.\n");
foreach my $line () {
if(! ($line =~ m/\>/)) {
chomp($line);
$proteinSeq .= $line;
}
}
close(fin);
# 每次都取 7 個 amino acid sequence, 然後換下一個位置
for(my $i = 0; $i < length($proteinSeq)-6 ; $i++) {
print "Pos ".($i+1).":";
# 將取出的amino acid sequence 存入 testSeq
my $testSeq = substr($proteinSeq,$i,7);
# 用來計算每個位置可能的 primer 總數
my $primerDesign = 1;
# 根據每條 aa sequence,依序拿出每一個 amino acid
for(my $j = 0; $j < 7; $j++) {
# 拿出每一個 amino acid
my $bp = substr($testSeq, $j, 1);
# 依照 codon table,取出相對應的 RNA sequence
my $codon = $reverse_code{$bp};
print "$codon";
# calculate degeneracy, 分析每個 RNA base 並計算所有 primer 數目
for(my $k = 0; $k < 3; $k++){
my $base = substr($codon, $k, 1);
# 若是 RYWMSK 其中一種,則有 2 個bps 可能
# 若是 VHDB 其中一種,有 3 種 bps 可能
# 若是 N, 有 4 種可能
if($base =~ m/[RYWMSK]/) { $primerDesign *= 2 }
elsif($base =~ m/[VHDB]/) { $primerDesign *= 3 }
elsif($base =~ m/[N]/) { $primerDesign *= 4 }
}
}
print "\t$primerDesign\n";
}
Reference
- Tore Samuelsson。Genomics and Bioinformatics。Cambridge university press。July 2012。ISBN: 9781107401242。