MSA on Evolution
Evolution: differences between humans and chimpanzees
With the complete genome sequencing of human and many other animals the next challenge is to figure out the similarities and differences in the genetic information between animals. For example, the human possesses advanced spoken, writing, abstract-level thinking and even music, painting, but these activities observed by the human in other animals are much less.
The significant differences between the human and the other animals are genes affecting brain functions.
But it seems not so distinct the genetic differences between the human and the chimpanzee. Between these two species, there are 35 million single nucleotide differences and 99% identical gene in the genome. About 5 millions insertions/deletions of nucleotide in the DNA, these occupy 3% in the genome.
A protein related to human speech
These differences in genes between animals imply the different functions.
- For example, there is a number of genomic regions conserved among the animals, but not in the human. These genes deleted may affect the regulation of gene expressions.
- Another example is the gene duplication event in humans, such as paralogous genes with new functions. As mentioned in another discussion previously(Link) the paralogous genes, ABL1 and ABL2, are the examples.
- Next example is about local changes, such as point mutations.
- These local changes may not only affect coding-gene sequences but also affect regulations of transcription or its process. These effects could give hints to learn about functions specific to humans, meaning the evolutions of different proteins in animals.
There are several proteins evolving specifically in the human lineage.
- non-coding RNA, HAR1F: this RNA involves in developing human neocortex which is a part of brain and is related with thought and language.
- genes encoding the protein alpha-tectorin: Based on the previous researches congenital deafness is related with mutations of alpha-tectorin-coding genes. This disease causes poor responses in the ear and that makes it difficult to understand the speech. The gene changes in alpha-tectorin in humans adjusted our hearing to develope human-specific speech.
- FOXP2 gene:
- FOXP2 is a transcription factor.
- FOXP2 protein exists in all vertebrates and its amino acid sequence is conserved.
- FOXP2 expressed in central nervous system (CNS) during the development.
- FOXP2 regulates a number of genes important for brain functions.
- FOXP2 is strongly associated with the human speech and language. When FOXP2 gene was mutated speech deficiency, difficulties with grammar, expressive language and defective articulation may be observed.
- Inherited defects related to FOXP2 gene are dominant, meaning both copies of a healthy FOXP2 gene are required for normal speech and language development.
- FOXP2 gene is located on human chromosome 7 and at least 3 isoforms exists.
FOXP2 genes in other animals
Most other vertebrates do not have advanced speech of the human but some vertebrates do present case in which mutations in the FOXP2 gene have the same effects with the human.
Based on the previous research mice that do not have a functional copy of FOXP2 are not able to produce the isolation calls. These calls are like young human babies crying in order to attract more attentions from their parents.
FOXP2 gene also plays a role in synaptic plasticity. This means connection between two neurons to be modulated. Such plasticity is important for the motor-skill learning. The motor-skill learning is associated with movement and activity.
FOXP2 gene is involved in neural development in birds. Experiments that the reduction of expression in FOXP2 to 50% in zebra finches shows they were not as efficient as normal birds imitating the songs taught.
Comparing FOXP2 in different animals
- Comparing FOXP2 sequences from a number of species is proceeded by multiple alignment.
- Search FOXP2(species) in a database, named swissport/UniProtKB, and collect the data searched in a file, named foxp2.fa.
- That more than two sequence alignment undergoes global alignment which is a pairwise alignment in a strict dynamic programming.
heuristic methods for the global alignment
- ClustalW: Thompson et al. (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics DOI: 10.1002/0471250953.bi0203s00
- Muscle: Edgar, R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5,113.
- T-coffee: Notredame et al. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol Biol 302(1),205-217
ClustalW construction
ClustalW
- progressive alignment procedures
- each step: a single sequence or sub-alignment is merged with a previous sequence
- The ordering in which sequence are aligned is following the guide tree. (Newick format, phylogenetic tree)
building and operation in linux commands:
# this command is to construct a database from swissprot in protein mode,
# the parameter, parse_seqids, is used for quick searching
$ makeblastdb -in ./swissprot -dbtype prot -parse_seqids
# find the amino acid sequence, named FOXP2_HUMAN,
# and capture the data into a file, named foxp2.fa
$ blastdbcmd -entry FOXP2_HUMAN -db ./swissprot > foxp2.fa
# find the another amino acid sequence, named FOXP2_MOUSE,
# capture the data and extend the content of the file, named foxp2.fa
$ blastdbcmd -entry FOXP2_MOUSE -db ./swissprot >> foxp2.fa
# ... (keep adding FOXP2-like sequences from different animals) ...
install ClustalW from [Link]
Opertions
# undergo global alignment of all the amino acid sequence saved in the file, named foxp2.fa,
# the parameter output=fasta is specific to outputting the fasta-mode file
# containing each sequence that is in a fixed length and that is seperated by a new line.
# besides that this process also produce two another files, named foxp2.dnd and foxp2.aln
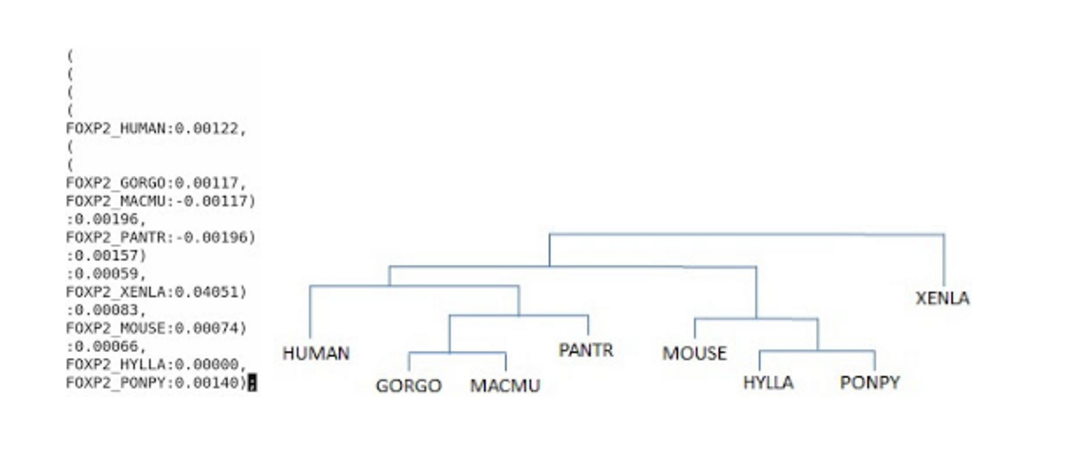
# foxp2.dnd saves results of the guide tree used as the ordering for multiple sequence alignment # foxp2.aln saves the whole seqence alignment results
$ ./clustalw2 ./foxp2.fa -output=fasta
- The guide tree with foxp2.dnd: the guide tree(right) is derived from the foxp2.dnd file(left)
- The implement is to find the amino acid whose position is in human and is different from all non-human amino acid.
#!/usr/bin/perl -w
use strict;
# variables declaration
my $human;
my $humanSeq;
my %nonHuman;
my $InputFileName = "foxp2.fasta";
my $flag = 0; # human or not
my $saveFlag = 0; # sequence saved or not
my $tempName = ""; # nonHuman sequence ID
my $tempSeq = ""; # nonHuman sequence
# read from file
open(fin,"$InputFileName") or die("File input error.\n");
foreach my $line () {
chomp($line);
if ($line =~ m/>/){
# save the seqNameID and its amino acid sequence into the hash
if($saveFlag) {
if(not $flag) { $nonHuman{$tempName}=$tempSeq; }
$tempSeq = "";
$saveFlag = 0;
}
# remove the first character of sequence ID
$line =~ s/>//;
if ($line =~ m/HUMAN/) {
$flag = 1;
$human = $line;
}
else {
$flag = 0;
$tempName = $line;
}
next;
}
if($flag) {
$humanSeq .= $line;
$saveFlag = 1;
}
else {
$tempSeq .= $line;
$saveFlag = 1;
}
}
# the final amino acid sequence must be saved into the hash
if($saveFlag) {
if(not $flag) { $nonHuman{$tempName}=$tempSeq; }
$tempSeq = "";
$saveFlag = 0;
}
close(fin);
# start to compare each site
# Try to find the amino acid whose position is in human and is different from all non-human amino acid
for(my $i = 0 ; $i < length($humanSeq); $i++) {
my $difference = 1;
foreach my $element (keys(%nonHuman)) {
if(substr($humanSeq, $i, 1) eq substr($nonHuman{$element}, $i, 1))
{
$difference = 0;
# in the worst condition, it is necessary to go throght all non-human amino acid sequence
# so as to check the position
last;
}
}
if($difference) {
print "pos:".($i+1)."\n";
print $human,"\t",substr($humanSeq,$i,1),"\n";
foreach my $element (keys(%nonHuman)) {
$tempSeq = $nonHuman{$element};
print "$element","\t",substr($tempSeq,$i,1),"\n";
}
}
}
- Execution result:

- Based on previous researches mice with mutations T303N(show 304 on the above image) & N325S seems affecting the basal ganglia on motor control & learning. The result does not contain N325S because the mutation N325S exists in frog Xenopus laevis.