Human disease cancer as a result of aberrant proteins
Background
The issue focuses on a aberration in the chromosome and this gives rise to a specific type of cancer.
There are more detailed researches and discussions in this moment, some references are following:
- Ludmil B. Alexandrov, Serena Nik-Zainal, David C. Wedge, Peter J. Campbel and Michael R. Stratton (2013) Deciphering Signatures of Mutational Processes Operative in Human Cancer. Cell Reports 3, 246–259
- Ludmil B. Alexandrov, Serena Nik-Zainal, David C. Wedge et al. (2013) Signatures of mutational processes in human cancer. Nature doi:10.1038/nature12477
Both the above researches that are continuous provide a new method to systematic classifications of ubiquitous cancer types.
Cancer as a genetics disease
- The cancer cell is characterized as a uncontrolled manner in its divisions. There are several reasons how this behavior achieves. First the overproduction of proteins that stimulate the cell growth. Second one is inactivation of the function that restricts cell growth. Third, the effects on cancer developing are from environment and lifestyle factors, such as tobacco and obesity. Forth one is the contributions of abnormal genes, such as the germ-line cell and the somatic cell. A well-known example of the aberrant germ-line gene that results in cancer is that abnormal BRCA1(or BRCA2) gene may elevate the risk of the breast cancer.
- There are lots of researches studying many types of cancer. One single mutation in gene is not enough to develop cancer. If there are lots of mutations in the same proteins-coding genes it would be more likely to develop the cancer. For example several mutations needed in well-studied colorectal cancer. These cancer cells originates from epithelial cells of the colon or rectum. The several mutations in genes related to Wnt signalling pathway in these cells may lead to the colorectal cancer. This pathway is important in embryogensis and cancer. One of the examples is APC (Adenomatosis ployposis coli).
- In cancer development it is often more than one gene mutated. A well-known example is mutation of TP53 gene which encodes the p53 protein. The p53 protein initiates cell death, apoptosis, when sensing a defect in Wnt signalling pathway. The mutation in this gene may deactivate the procedures of cell death in abnormal Wnt signalling pathway.
- Besides TP53 gene there are other genes that their functions in apoptosis and that is deactivated in colorectal cancer, such as the genes encoding TGF-beta and DCC(Deleted in colorectal cancer).
The genes that is mutated and that is implicated in cancer are sometimes classified into two groups.
- Oncogene: It is a gene when mutated will promote cancer develop.
- Tumor suppressor gene: It is a gene that is normally prevent the cell from critical steps towards cancer. When a mutation in tumor suppressor gene it would be prone to developing cancer.
Cancer & DNA repair
- Under exposures to chemical mutagens and UV radiation erroneous base sequences or double-strand breaks are often found in the DNA molecules. There are complex biological mechanisms for repairing DNA damages in all kinds of cell.
- There are some types of cancer related to the gene mutations in repairing DNA molecular. For example, Xeroderma Pigmentosum is a inherited mutations in genes involved in nucleotide excision repair (NER). When the NER mechanism is not functional it is prone to developing skin cancers. The UV radiation may cause DNA damages from formations of pyrimidine dimer in skin cells. This is the cross-linking of two adjacent pyrimidines in one DNA strand and normally these DNA damages are removed by NER.
- Another example of disease related with DNA repair is Lynch Syndrome. The disease is caused by the mutations in two genes, MSH2 and MLH1, involved in DNA mismatch repairs. Normally the DNA mismatch repair identifies and corrects errors in a newly synthesized strand of DNA.
Another example of disease is Li-Fraumeni Syndrome. There are variety of different cancer types, such as breast cancer and brain tumors, in the patients with this disease. This disease is caused by mutations in genes coding p53 protein. The p53 protein is
- the control protein of cell division and growth.
- making sure that DNA molecule is adequately repaired before proceeding to synthesis of new DNA by growth arrest at a point in the cell cycle referred to as the G1/S checkpoint.
Chromosome rearrangements and the Philadelphia chromosome
Chromosomes have another character that involves more extensive rearrangements of genetic materials.
- homologous recombination : For example, this takes place in germ-line cells during crossing-over in meiosis. Two homologous chromosomes line up and the segments of chromosomes are changed.
- chromosomal trans-location : Genetic materials are changed between non-homologous chromosomal regions. A number of mechanisms may operate to result in such trans-location, such as homologous recombination.
Chromosomal trans-location is typically harmless. But there are some cases that result from such rearrangements of cancer diseases. Fusion proteins are often produced in such diseases. The fusion proteins are formed as a result that gene segments are combined so as to produce an aberrant gene product from two different genes. The fusion proteins are sometimes connected with diseases.
- c-myc (chr8) / IGH (chr14) → burkitt's lymphoma
- PAX8 (chr2) / PPAR-gamma one (chr3) → follicular thyroid cancer
- JAK (chr9) / TEL (chr12) → chronic myelogenous leukaemia (CML) and acute lymphoblastic leukaemia (ALL)
- BCR (chr9) / ABL (chr22) → CML and ALL
CML gives rise to a very high production of WBC.
BCR/ABL is a fusion protein consisting of gene materials from chr9 and chr22, refer to the following image
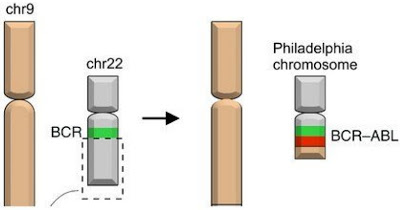
- Both chromosome 9 and 22 are identified by a microscope.
- After trans-location chromosome 22 is much smaller then its normal counterpart.
- This fusion protein was discovered in 1960 by Peter Nowell and David Hungerford at facilities located in the city of Philadelphia.
- The function of BCR is still unknown.
- On the contrary, ABL includes two types, ABL1 and ABL2. ABL1 is involved in BCR-ABL fusion process. ABL1 has tyrosine kinase activities and it is helpful for cell differentiation and cell division.
- The fusion between BCR and ABL protein results in an activation of tyrosine kinase of ABL. This activation leads the cells to the state of uncontrolled cell growth.
- The breakpoint for funsion between BCR and ABL is not always the same. There are many fusion proteins in different forms.
Next section would take one of the BCR/ABL forms into considerations detailed.
Dotplots and alignments
There is a concept, named sequence alignment, to compare BCR-ABL fusion protein with the normal BCR and normal ABL protein. The sequence alignment tries to fit the sequence optinally to each other.
When it comes to talk about the sequence alignment a 2-D plot, named dotplot, is an access point. The following is an example.
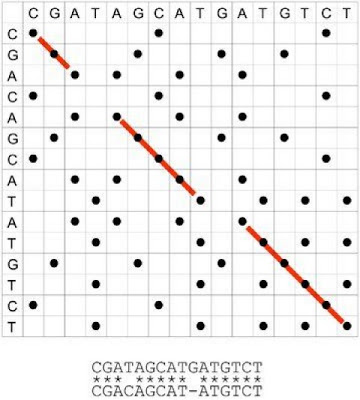
- The value on the horizontal axis is one sequence and the value on the vertical axis is the other sequence.
- A dot represents a match of nucleotides between two sequences.
- After the compare three different diagonals are combined to produce the alignment at the bottom.
- In order to accommodate the matches in the alignment a gap is introduced into the sequences, represented by dash (-). This gap means in biological process mutation events. For example, a part of nucleotides in the sequence have been inserted and deleted.
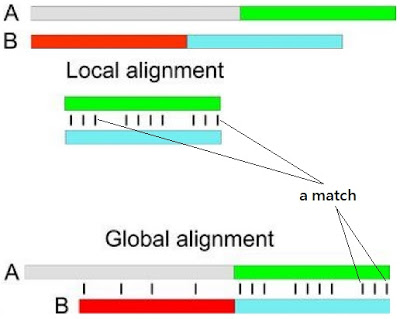
There are more better methods to produce an optimal alignment. There are two different types of alignment based on the overall or partial sequence.
- global alignment : Two sequences in their full length are optimally aligned.
- local alignment : Only regions of significant similarity between the sequences are reported.
The bottom image is the difference between the global and local alignment.
There is a algorithm, named dynamic programming, suitable for dealing with the local alignments. The concept of dynamic programming is referred as the following image.

- The scoring scheme in the dynamic programming is divided into three elements, matches, dis-matches and gap penalties. In this example assume the score of a match is 2, the score of a dis-match is -1 and gap penalty is -2.
- Two sequences, TAAAA and CAACAA, are used for this example.
- The two index, i and j, represent the increment pointers of nucleotides in TAAAA and CAACAA sequence.
- 1st step: the 2-D matrix(F), the value of F(i,0) and F(0,j) are set to zero.
- 2nd and further step: The other value of F(i,j) are built recursively based on the rule of panel A in the above image. For example F(1,1) has two values at first, -1 and -2. The score of -1 means a dis-match between C and T. The score of -2 means a gap between C and "(None)" or a gap between T and "(None)". Because the negative value is not needed in finding the local alignment the value of F(1,1) is set to zero. (The reason why penal A exists a option that the value is zero.)
- To produce the final alignment it is a good starting from the highest value in the matrix(F). Trace backwards to calculate which conditions(match, dis-match or gap) are suitable for this position. In the example on the above image the red arrow indicates the direction from the beginning to the end (F(i,j) = 0).
Unlike the nucleotides not all amino acid replacements take place in the same frequency, so it is necessary to take a distinct property of protein evolution into considerations. For example the replacement from glutamic acid to aspartic acid is much more common than the replacement from glutamic acid to cysteine. The explanation of these differences is partly related to the physical and chemical differences between amino acids. The glutamic acid and aspartic acid are both negatively charged and replacement between these two amino acids may not influence the structure and function a lot.
The frequency of replacement between two different amino acids may be exploited in the scoring of alignments. A likely replacement receives a higher score than an unlikely one. An amino acid substitution matrix, such as BLOSUM62, is a scoring table representing the probability of the particular amino acid substitution. The following is the BLOSUM62 table.

References
- Genomics and Bioinformatics。Tore Samuelsson。Cambridge university press。July 2012。ISBN: 9781107401242。