DP-bind
Service
- Web server: http://lcg.rit.albany.edu/dp-bind/
Example
- protein FC-FOS (PDB: 1A02)
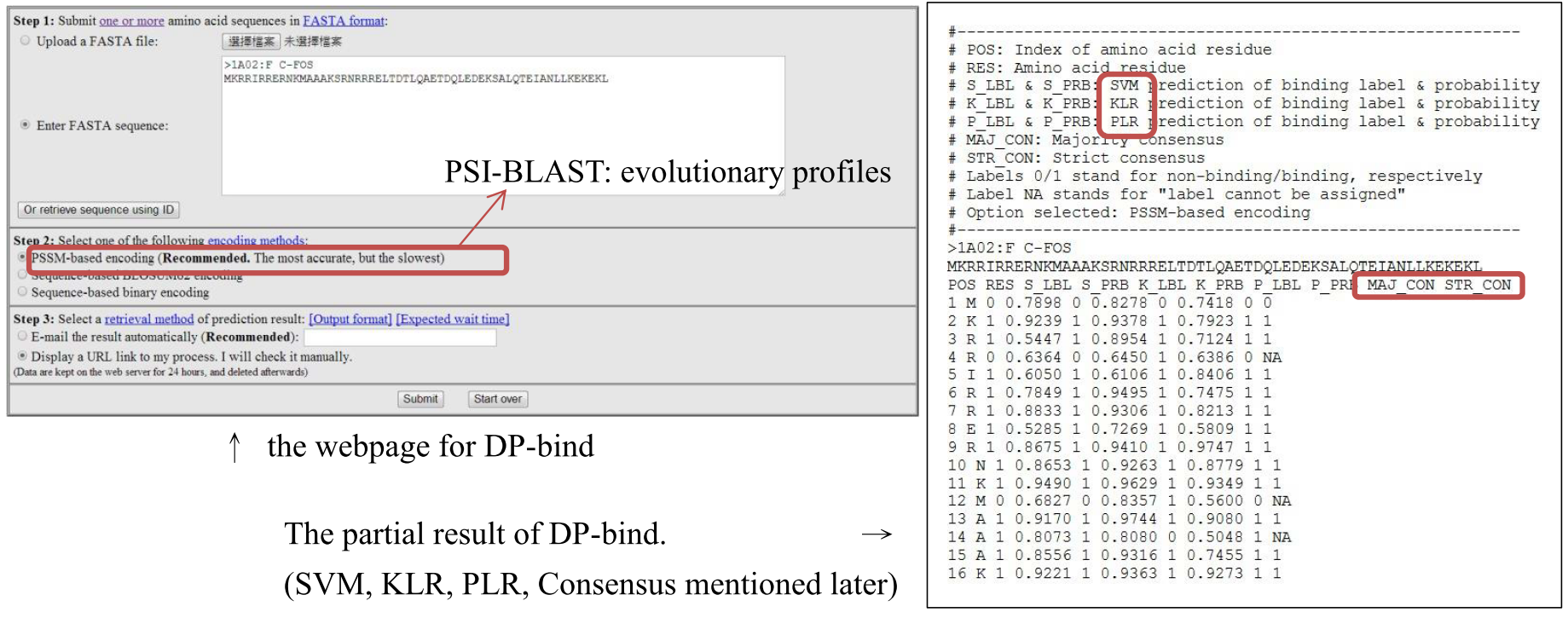
Classifier and Major method
Two encoding methods for transforming amino acids into numeric format for calculating, that are PSSM-based and sequence-based (divided into BLOSUM62 and binary encoding)
Three kinds of the classifier: These three methods are classified into supervised pattern recognition in the machine-learning.
- Support vector machine (SVM): developed by Vapnik in 1998
- Kernel logistic regression (KLR): developed by Zhu and Hastie in 2005
- Penalized logistic regression (PLR): developed by le Cessie and van Houwelingen in 1992
Combination among results: Results from 3 classifiers are combined as one consensus result.
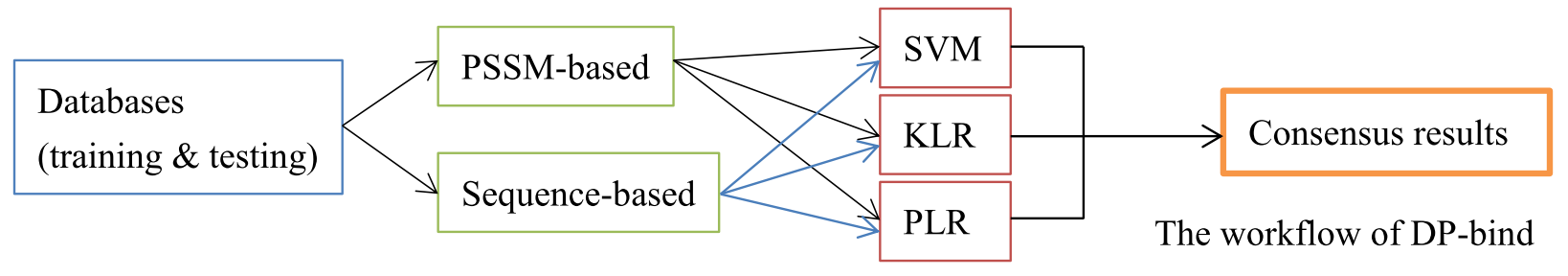
Dataset
PDNA-62: This dataset consisted of 62 protein sequences (non-redundant) which are experimentally solved protein-DNA complexes, utilized many time in the previous studies [5] and downloaded from the Protein Data Bank (PDB).
The training and testing procedures: Test the predictors through leave-out-protein-out cross validation.
Results
The best single prediction combination: PSSM-based and KLR in this dataset.
The consensus result would be the best prediction of the DNA-binding residues. Three classifiers represent the three aspects of biological meanings and considerations so that the performance would be the best one.
The major and strict consensuses are determined by voting from three classifiers.

Independent dataset
- There is no independent dataset tested in this paper.
Reference
Jagat S Chauhan et al. (2009) Identification of ATP binding residues of a protein from its primary sequence. BMC Bioinformatics 10:434 doi:10.1186/1471-2105-10-434
Lukasz Kurgan et al. (2011) ATPsite: sequence-based prediction of ATP-binding residues. Proteome Science 9(Suppl 1):S4
Manning, G., Whyte, D.B., Martinez, R., Hunter, T. and Sudarsanam, S. (2002) The protein kinase complement of the human genome. Science 298, 1912-1934
Zhou FF, Xue Y, Chen GL, Yao X. (2004) GPS: a novel group-based phosphorylation predicting and scoring method. Biochem Biophys Res Commun 24;325(4):1443-8.
Shandar Ahmad, M. Michael Gromiha and Akinori Sarai (2004) Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. BIOINFORMATICS Vol. 20 no. 4, pages 477–486 DOI: 10.1093/bioinformatics/btg432
Seungwoo Hwang, Zhenkun Gou and Igor B. Kuznetsov(2007) DP-Bind: a web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. BIOINFORMATICS Vol. 23 no. 5, pages 634–636 doi:10.1093/bioinformatics/btl672